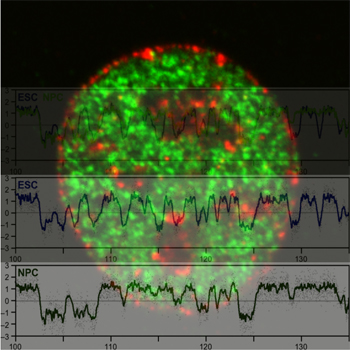
the sizes and locations of early (green) and late (red)
replicating chromosome segments and demonstrate that
they are highly conserved within a cell type, but globally re-organized and consolidated into larger coordinately
replicating segments during differentiation of embryonic
stem cells. (Photo: FSU Department of Biological Science)
In a groundbreaking study led by an eminent molecular biologist at Florida State University, researchers have discovered that as embryonic stem cells turn into different cell types, there are dramatic corresponding changes to the order in which DNA is replicated and reorganized.
The findings bridge a critical knowledge gap for stem cell biologists, enabling them to better understand the enormously complex process by which DNA is repackaged during differentiation — when embryonic stem cells, jacks of all cellular trades, lose their anything-goes attitude and become masters of specialized functions.
As a result, scientists now are one significant step closer to the central goal of stem cell therapy, which is to successfully convert adult tissue back to an embryo-like state so that it can be used to regenerate or replace damaged tissue. Such therapies hold out hope of treatments or cures for cancer, Parkinson’s disease, multiple sclerosis, spinal cord injuries and a host of other devastating disorders.
Using mouse and human embryonic stem cells, FSU researchers employed advanced imaging techniques and state-of-the-art genomics technology to demonstrate, with unprecedented resolution along long stretches of chromosomes, which sequences are replicated first, and which occur later in the process of differentiation.
“Understanding how replication works during embryonic stem cell differentiation gives us a molecular handle on how information is packaged in different types of cells in manners characteristic to each cell type,” said David M. Gilbert, the study’s principal investigator. “That handle will help us reverse the process in order to engineer different types of cells for use in disease therapies.” Internationally renowned for his body of cutting-edge research on chromosomal structure and reproduction that he began as a doctoral student at Stanford University in the 1980s, Gilbert joined the FSU faculty and was appointed as the first J. Herbert Taylor Distinguished Professor of Molecular Biology in 2006.

Results from the FSU study, which includes contributions from researchers at three other institutions, are described in a paper published in the October 7, 2008, edition of PLoS (Public Library of Science) Biology, a peer-reviewed journal that showcases biological science research of exceptional significance. So prodigious were the findings that the current paper — “Global Reorganization of Replication Domains During Embryonic Stem Cell Differentiation” — is focused solely on results observed in the mouse embryonic stems cells; data on the human cells will be detailed in a future report.
“We know that all the information (DNA) required to take on the identity of any tissue type is present in every cell, because we already can, albeit very inefficiently, create whole animals from adult tissue through cloning,” Gilbert said. “We also can make a kind of artificial embryonic stem cells, called induced pluripotent stem cells, out of many adult cell types, but there are two major hurdles remaining. First, the methods currently used rely on the unnatural retroviral insertion of genes into patients’ cells, and these genes are capable of forming tumors. Second, this method is very inefficient as well because only one in 1,000 cells into which the genes are inserted becomes pluripotent. We must learn how cells lose pluripotency in the first place so we can do a better job of reversing the process without risks to patients.
“The challenge is, adult cells are highly specialized and over the course of their family history over many generations they’ve made decisions to be certain cell types rather than others,” he said. “In doing so, they have tucked away the information they no longer need on how to become other cell types. Hence, all cells contain the same genetic information in their DNA, but during differentiation they package it with proteins into ‘chromatin’ in characteristic ways that define each cell type. The rules that determine how cells package DNA are complicated and have been difficult for scientists to decipher.”
But, Gilbert noted, one time that the cell “shows its cards” is during DNA replication.
“During this process, which was the focus of our FSU research, it’s not just the DNA that replicates,” he said. “All the packaging must be replicated as well in each cell division cycle.”
He explained that embryonic stem cells have many more, smaller “domains” of organization than differentiated cells, and it is during differentiation that they consolidate information.
“In fact, ‘domain consolidation’ is what we call the novel concept we discovered,” he said.
Gilbert likened the concept of domain consolidation to the undeclared or “undifferentiated” college student who then consolidates her literature resources during the course of declaring a major and specialization. “From a student with books on all subjects on all of her bookshelves comes a student who has placed all texts pertaining to her major on the eye-level shelf and moved the distantly-related, potentially distracting texts to the hard-to-reach bottom or top shelves,” he said.
“Now, our challenge as scientists,” said Gilbert, “is to build on what we’ve learned about domain consolidation so that we can efficiently and safely create patient-specific induced pluripotent stem cells or even coax the body’s cells to change their specialization in response to medications.”
Funding for the study came largely from grants awarded to Gilbert from the National Institutes of Health. To access a copy of the paper on the PLoS Biology Web site, visit this link. Learn more about Gilbert’s research achievements at gilbertlab.bio.fsu.edu.
Paper co-authors from the FSU Department of Biological Science include postdoctoral assistants Ichiro Hiratani and Tomoki Yokochi and doctoral student Tyrone Ryba. Contributions also came from researchers at the Friedrich Miescher Institute for Biomedical Research in Basel, Switzerland; the University of Alabama at Birmingham Schools of Medicine and Dentistry; and the State University of New York, Upstate Medical University, Syracuse.




